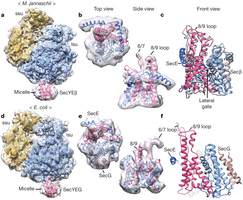
Dear School of Physics Community,
Please join me congratulating Claire Berger, Chandra Raman, Elisa Riedo, Mike Schatz and Deirdre Shoemaker. They have been named Fellows of the American Physical Society (citations below). APS Fellowship is a recognition by their peers of their exceptional contributions to physics. This is a distinct honor as the number of Fellows of the APS is limited to no more than one half of one percent of the membership.
CITATIONS:
Claire Berger (Division of Material Physics) “For seminal contributions to the development of epitaxial graphene electronics”
Mike Schatz (Topical Group on Statistical and Nonlinear Physics) “For pioneering and creative experimental contributions to the characterization and control of complex fluid and pattern formation phenomena”
Deirdre Shoemaker (Division of Computational Physics) “For her leading role in the investigation of dynamical and black-hole space-times and their observational signatures”
Elisa Riedo (Division of Condensed Matter Physics) "For atomic force microscopy studies of nanoscale friction, liquid structure and nanoscale elasticity, and the invention of thermochemical nanolithography.”
NSF CAREER Proposal
Shina Tan’s NSF CAREER proposal on "Few-body and many-body theory of ultracold atoms and molecules” has been recommended for funding.